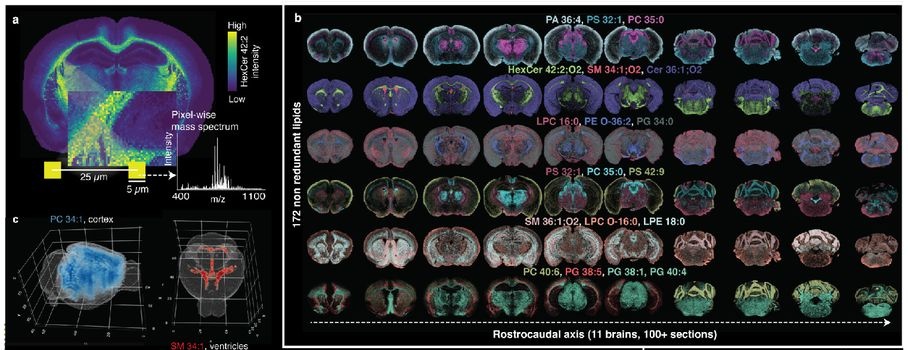
The first comprehensive lipidomic atlas of the adult mouse brain, mapping 172 membrane lipids and identifying 539 spatially coherent biochemical domains (lipizones).
Fusar Bassini L, Schede HH, Capolupo L, Alieh LHA, Venturi F, Valente A, Droin C, Trejo Banos D, Khven I, Asirim EZ, Nasrallah A, Kaysudu I, Krymova E, D’Angelo G, La Manno G
bioRxiv,
2025
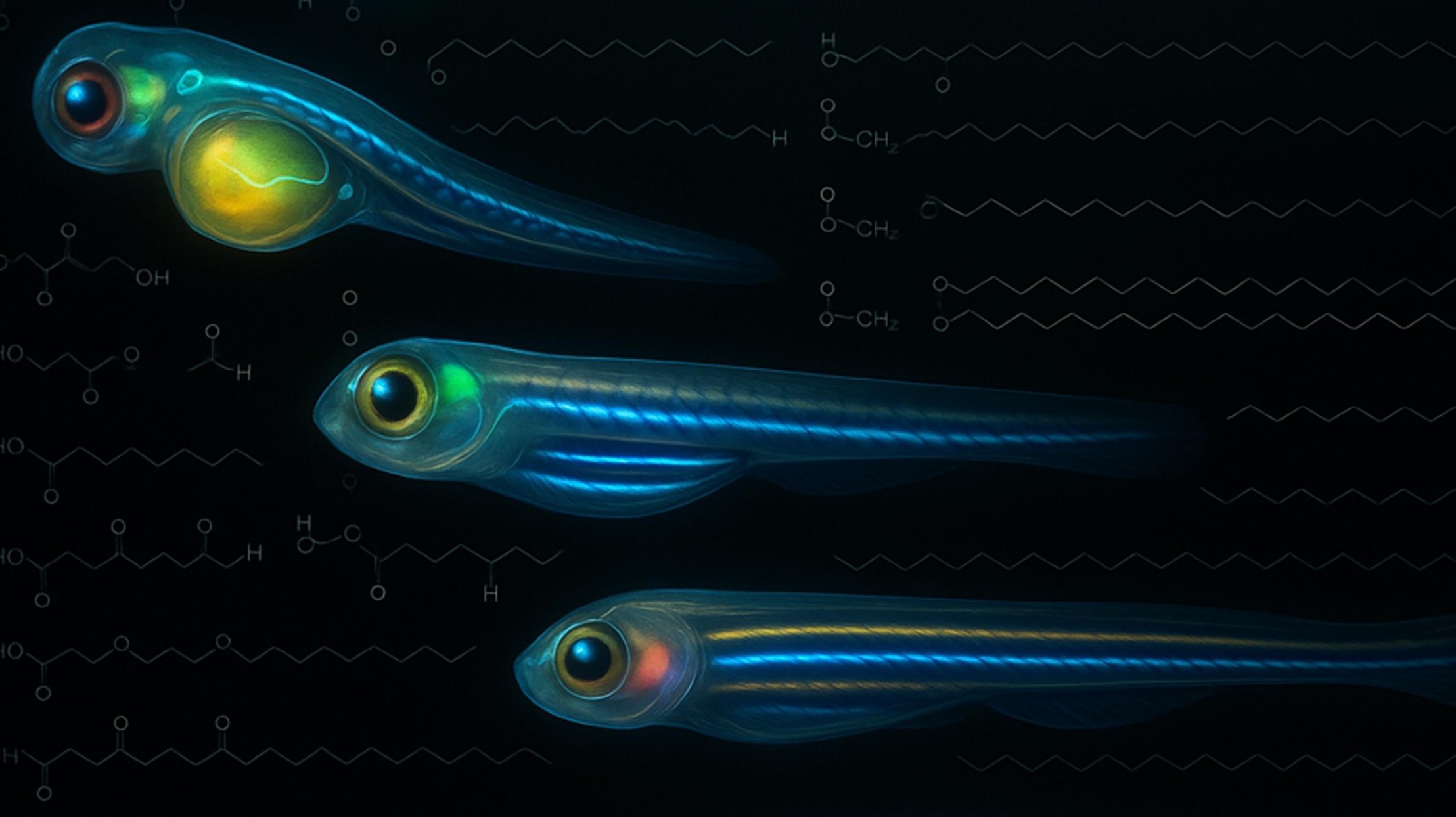
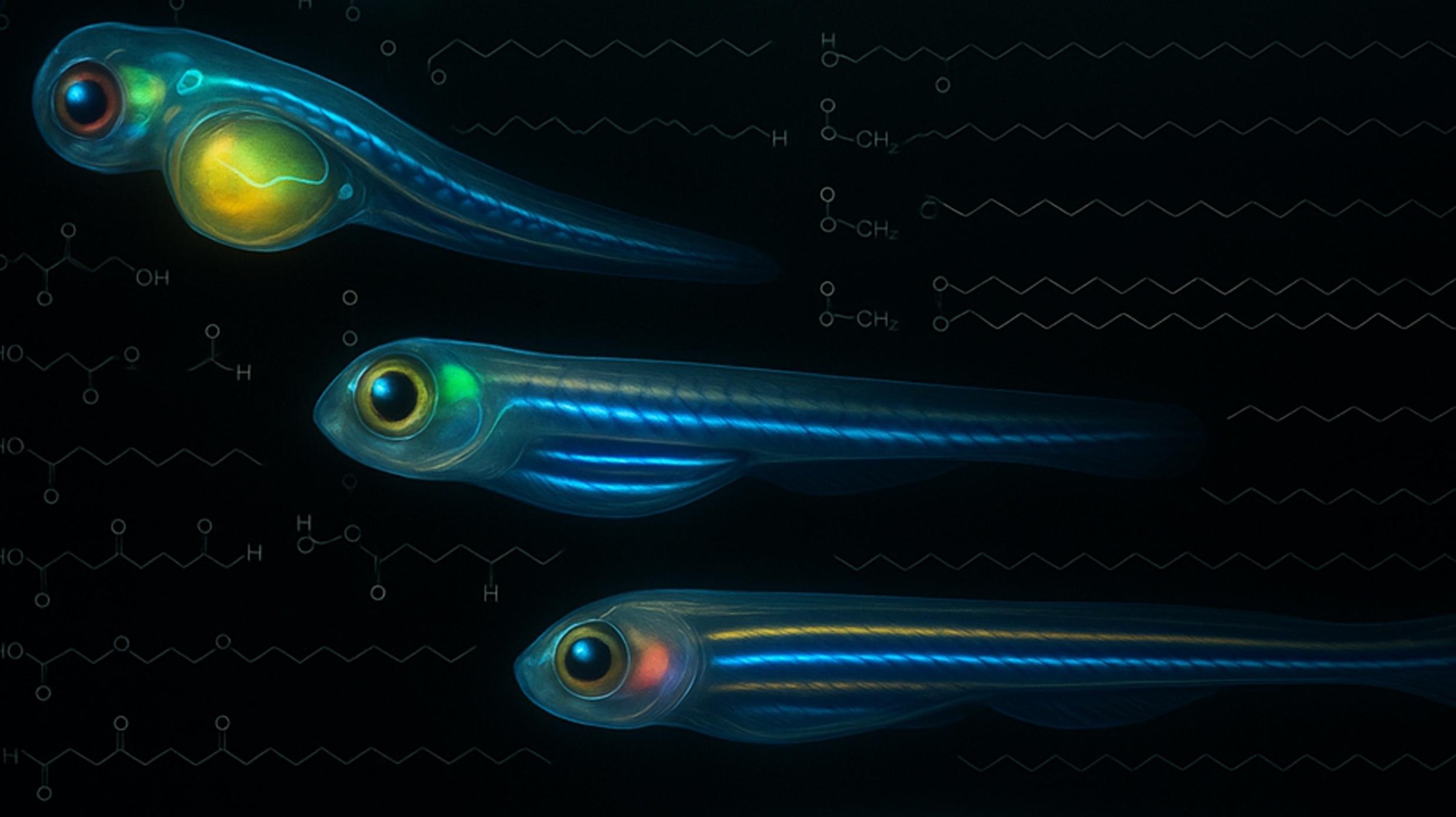
We present uMAIA, an analytical framework for spatial lipidomics, and use it to map the 4D distribution of over a hundred lipids in the developing zebrafish embryo, revealing metabolic trajectories that unfold in concert with morphogenesis.
Halima H. Schede, Leila H. A. Alieh, Laurie A. Rohde, Antonio Herrera, Anjalie Schlaeppi, Guillaume Valentin, Alireza Gargoorimothlag, Albert Dominguez Mantes, Celine Jollivet, Julijana Paz-Montoya, Lucile Capolupo, Irina Khven, Andrew C. Oates, Giovanni D’Angelo, Gioele La Manno
Nature Methods,
2025
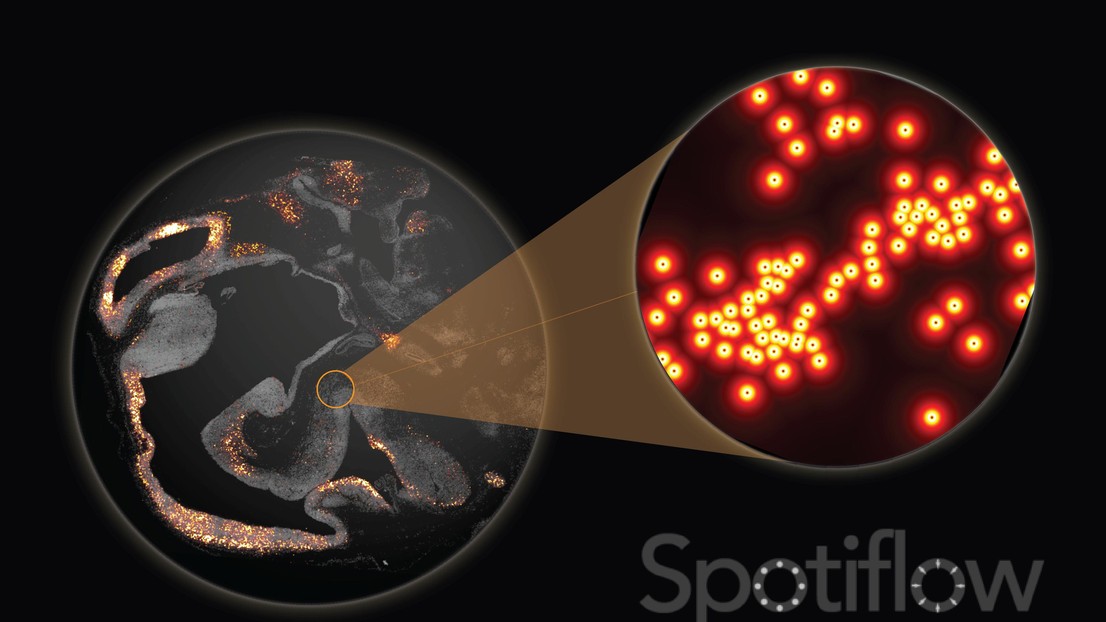
Spotiflow is a deep learning method for subpixel-accurate spot detection in fluorescence microscopy that achieves state-of-the-art performance while being robust to different noise conditions.
Albert Dominguez Mantes, Antonio Herrera, Irina Khven, Anjalie Schlaeppi, Eftychia Kyriacou, Georgios Tsissios, Evangelia Skoufa, Luca Santangeli, Ekaterina Buglakova, Emine Berna Durmus, Suliana Manley, Anna Kreshuk, Detlev Arendt, Can Aztekin, Joachim Lingner, Gioele La Manno, Martin Weigert
Nature Methods,
2025
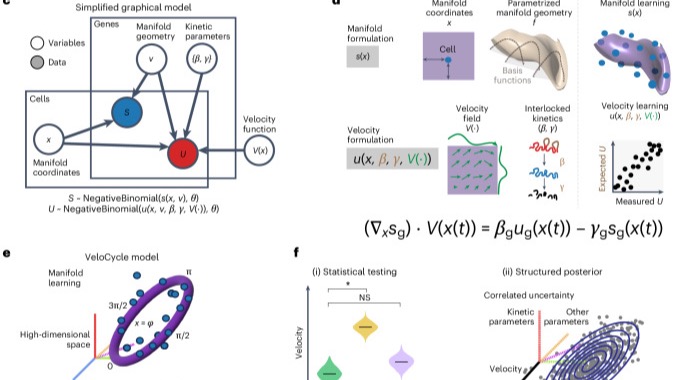
VeloCycle is a Bayesian RNA velocity model enabling formal statistical testing of gene expression dynamics in single-cell data, used to identify developmental modulations of the cell cycle.
Alex R. Lederer, Maxime Leonardi, Lorenzo Talamanca, Antonio Herrera, Colas Droin, Irina Khven, Hugo J. F. Carvalho, Alessandro Valente, Albert Dominguez Mantes, Pau Mulet Arabí, Luca Pinello, Felix Naef, Gioele La Manno
Nature Methods,
2024
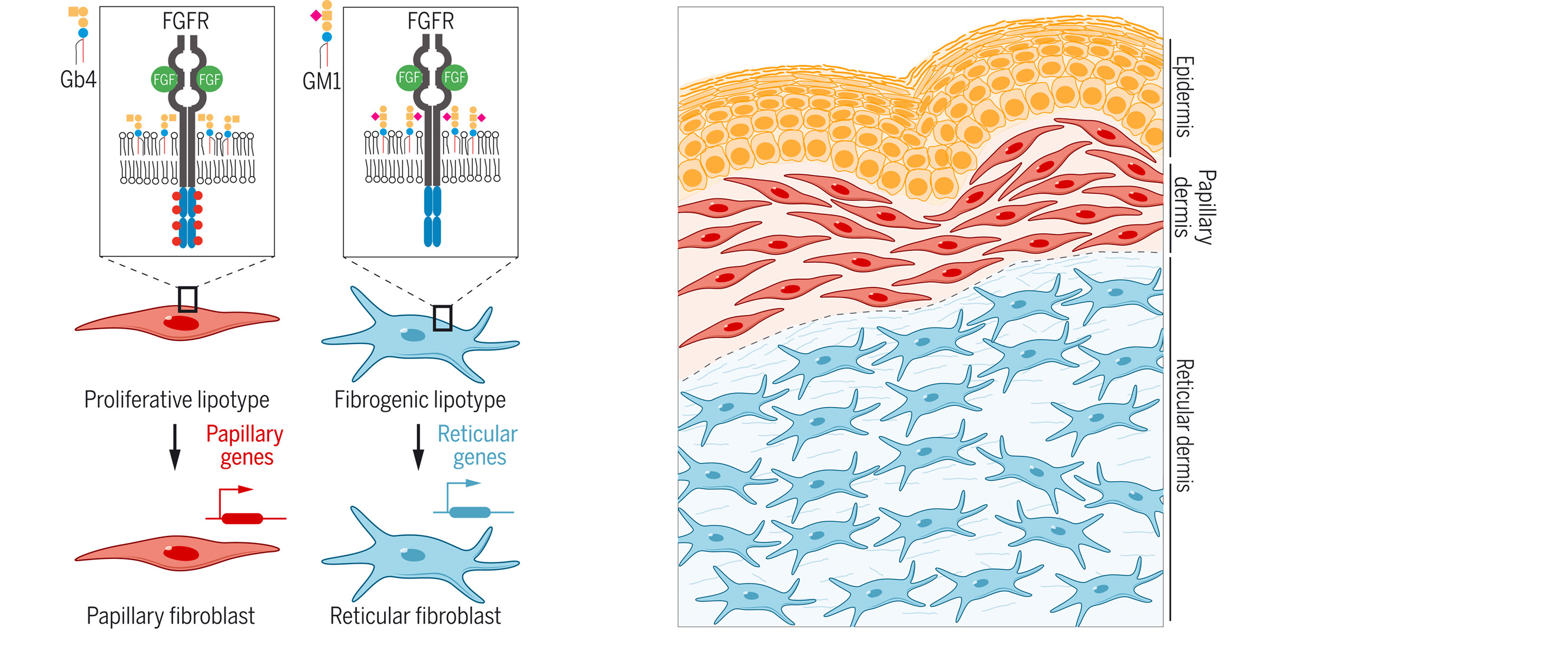
Cell-to-cell lipid heterogeneity affects the determination of cell states, adding a new regulatory component to the self-organization of multicellular systems.
Capolupo L, Khven I, Lederer AR, Mazzeo L, Glousker G, Ho S, Russo F, Montoya JP, Bhandari DR, Bowman AP, Ellis SR, Guiet R, Burri O, Detzner J, Muthing J, Homicsko K, Kuonen F, Gilliet M, Spengler B, Heeren RMA, Dotto GP, La Manno G, D’Angelo G
Science,
2022
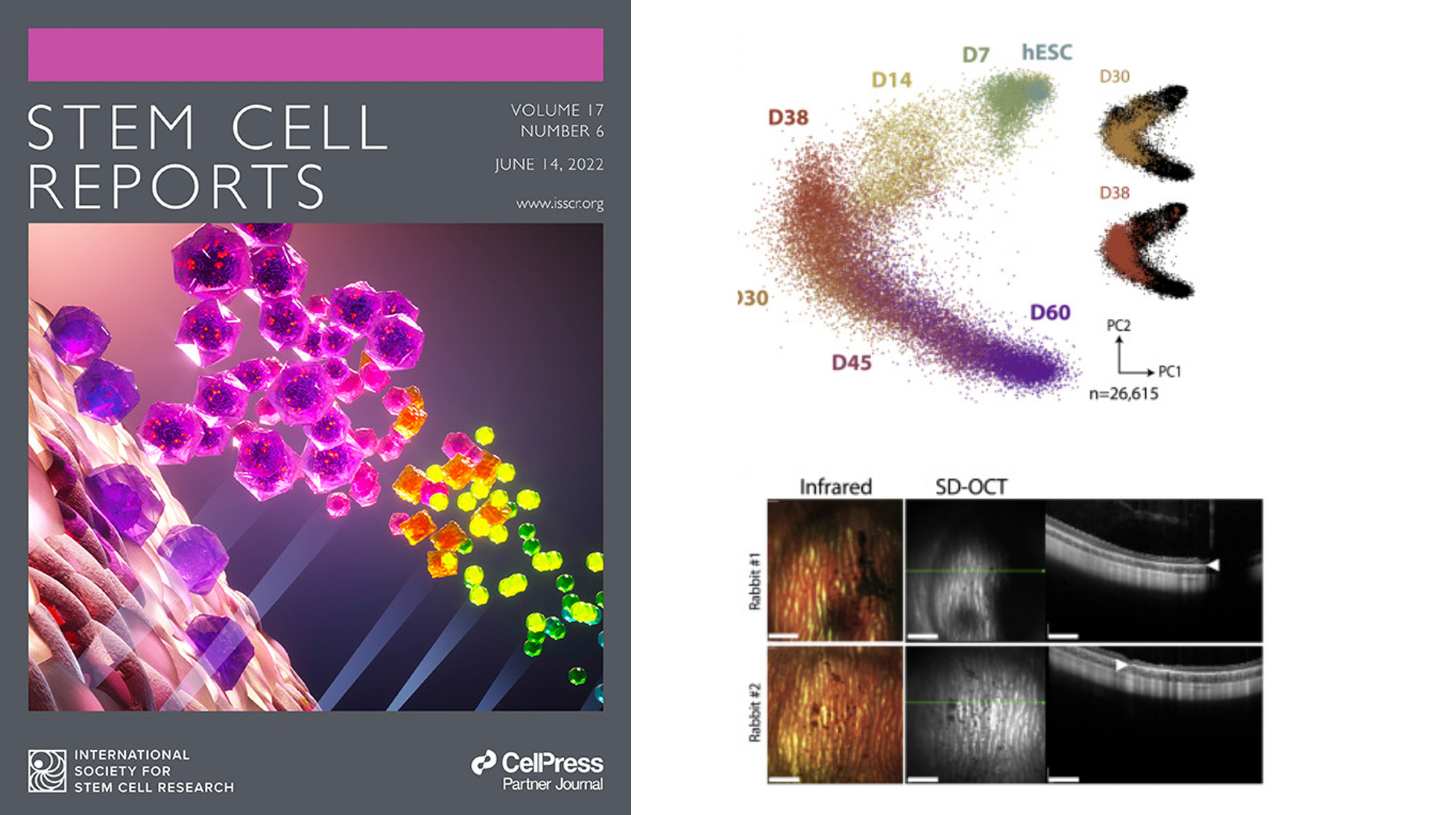
Human embryonic stem cell-derived retinal pigment epithelial cells (hESC-RPE) are studied as potential treatment for age-related macular degeneration (AMD). The study conducts single-cell transcriptomic profiling of an hESC-RPE differentiation protocol developed for clinical use, demonstrating its ability to produce a pure RPE pool.
Sandra Petrus-Reurer, Alex R Lederer, Laura Baqué-Vidal, Iyadh Douagi, Belinda Pannagel, Irina Khven, Monica Aronsson, Hammurabi Bartuma, Magdalena Wagner, Andreas Wrona, Paschalis Efstathopoulos, Elham Jaberi, Hanni Willenbrock, Yutaka Shimizu, J Carlos Villaescusa, Helder André, Erik Sundstrӧm, Aparna Bhaduri, Arnold Kriegstein, Anders Kvanta, Gioele La Manno, Fredrik Lanner
Stem Cell Reports,
2022
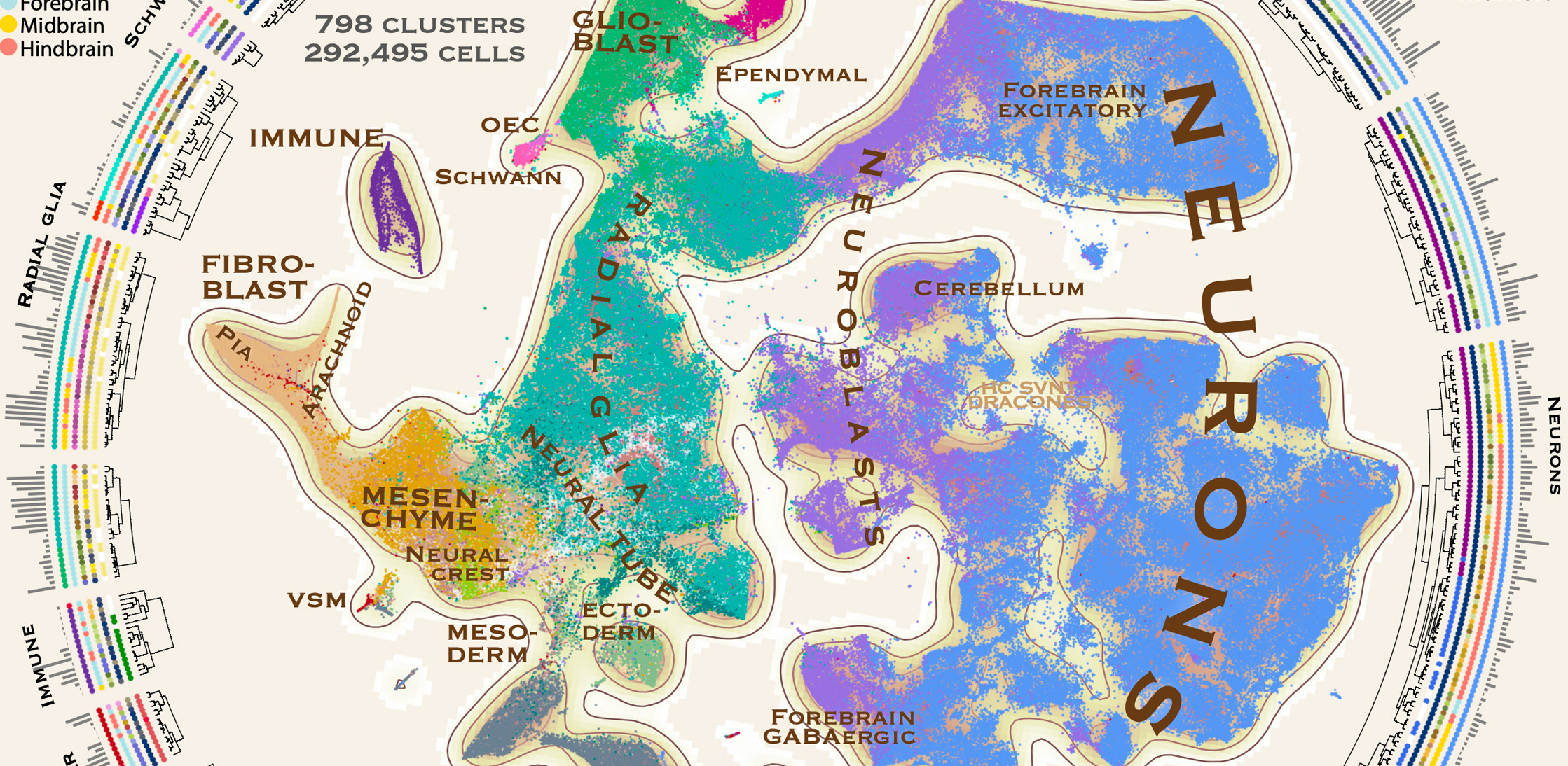
We report a comprehensive single-cell transcriptomic atlas of the embryonic mouse brain between gastrulation and birth. We identified almost eight hundred cellular states that describe a developmental program for the functional elements of the brain and its enclosing membranes.
Gioele La Manno, Kimberly Siletti, Alessandro Furlan, Daniel Gyllborg, Elin Vinsland, Alejandro Mossi Albiach, Christoffer Mattsson Langseth, Irina Khven, Alex R. Lederer, Lisa M. Dratva, Anna Johnsson, Mats Nilsson, Peter Lönnerberg, Sten Linnarsson
Nature,
2021
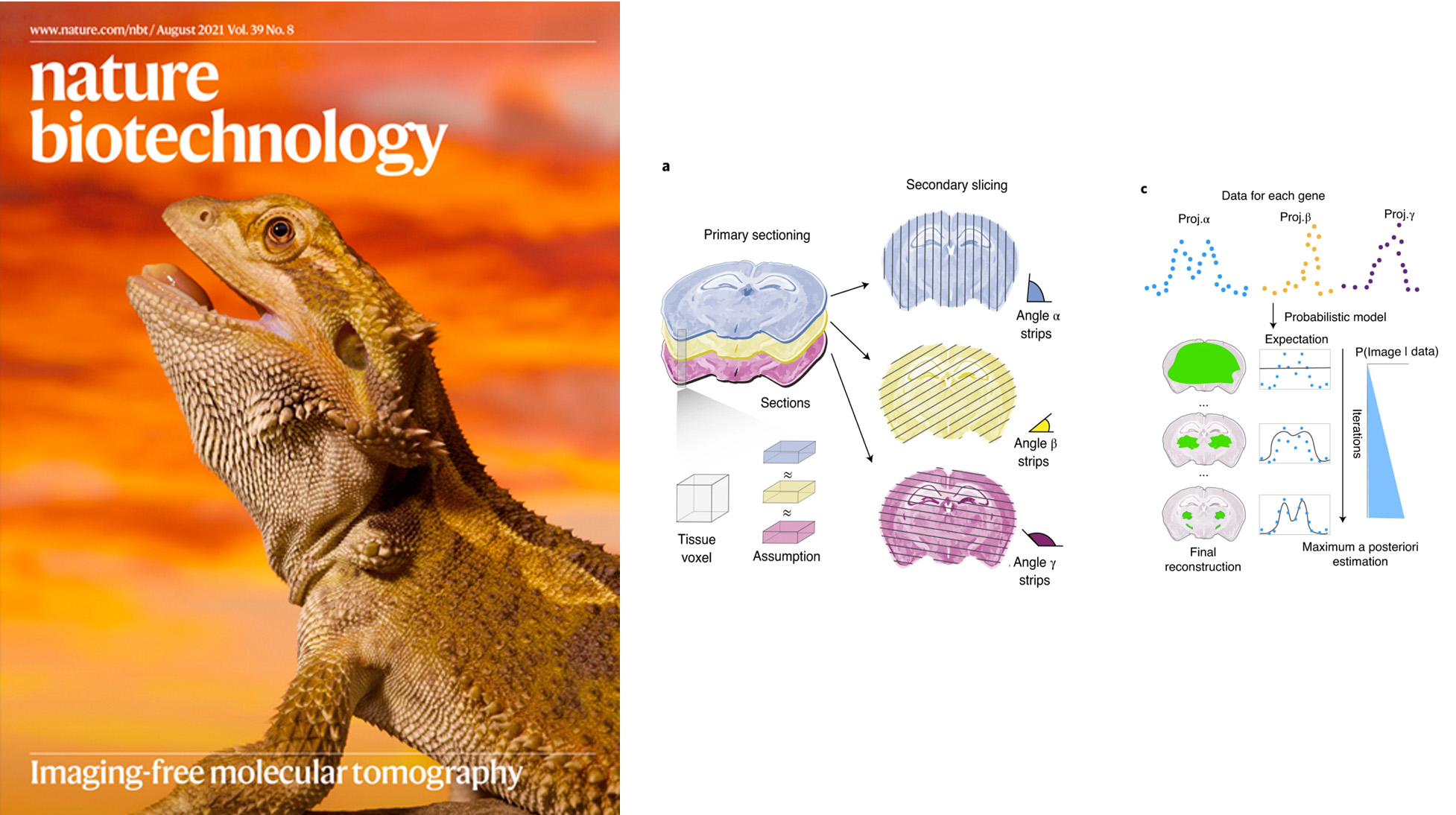
We describe an imaging-free framework to localize high-throughput readouts within a tissue by cutting the sample into thin strips in a way that allows subsequent image reconstruction. We applied the technique to the brain of the Australian bearded dragon, Pogona vitticeps. Our results reveal the molecular anatomy of the telencephalon of this lizard, providing evidence for a marked regionalization of the reptilian pallium and subpallium.
Halima Hannah Schede, Christian G Schneider, Johanna Stergiadou, Lars E Borm, Anurag Ranjak, Tracy M Yamawaki, Fabrice P A David, Peter Lönnerberg, Maria Antonietta Tosches, Simone Codeluppi, Gioele La Manno
Nature Biotechnology,
2021
Here we show that RNA velocity - the time derivative of RNA abundance - can be estimated by distinguishing unspliced and spliced mRNAs in standard single-cell RNA sequencing protocols. RNA velocity is a vector that predicts the future state of individual cells on a timescale of hours.
Gioele La Manno, Ruslan Soldatov, Amit Zeisel, Emelie Braun, Hannah Hochgerner, Viktor Petukhov, Katja Lidschreiber, Maria E. Kastriti, Peter Lönnerberg, Alessandro Furlan, Jean Fan, Lars E. Borm, Zehua Liu, David van Bruggen, Jimin Guo, Xiaoling He, Roger Barker, Erik Sundström, Gonçalo Castelo-Branco, Patrick Cramer, Igor Adameyko, Sten Linnarsson & Peter V. Kharchenko
Nature,
2018